Abstract

Drs. Armistead and Vincent are co-last authors.
Background
T cell responses to minor histocompatibility antigens (mHAs) mediate graft versus leukemia (GvL) effects and graft versus host disease (GvHD) after allogeneic hematopoietic cell transplantation (alloHCT). Therapies that boost T cell responses improve the efficacy of alloHCT; however, these have been limited by concurrent increases in incidence and severity of GvHD. mHAs with expression restricted to hematopoietic tissue (GvL mHAs) are attractive targets for driving GvL without increasing GvHD risk. Prior work to identify mHAs has focused on small sets of mHAs or population-level SNP association studies (Oostvogels et al 2016, Martin et al 2017). We report here an approach to identify GvL mHAs shared across many donor-recipient pairs (DRPs) within a large population, particularly for HLA alleles common in different US genomic ancestry groups. We used genotyping data from the DISCOVeRY-BMT (Determining the Influence of Susceptibility Conveying Variants Related to one-Year mortality after BMT) dataset of 3233 HLA-matched alloHCT DRPs to predict novel GvL-associated mHAs, determined the number of predicted mHAs needed to cover all recipients with the given HLA alleles, and confirmed presentation of 24 novel predicted GvL mHAs using mass spectrometry (MS).
Methods
DRP genotyping and clinical data were derived from the DISCOVeRY-BMT study, including Acute Myeloid Leukemia (AML), Acute Lymphocytic Leukemia (ALL), and Myelodysplastic Syndrome (MDS) patients treated with alloHCT and reported to CIBMTR 2000-2011. Genotyping was performed using the Illumina human OmniExpress 1.0 array. Minor mismatches were defined as SNP loci where the graft recipient contained an allele absent in the donor, and mHAs were peptides derived from the recipient allele predicted to bind recipient HLA. We then labeled mHAs as "GvL," "GvH," or "both" based on mHA source gene tissue expression. We applied a custom greedy algorithm solution to the maximum set coverage problem to generate ranked lists of the smallest sets of GvL mHAs needed to cover 100% of DRPs with each HLA allele. We then used three AML cell lines expressing HLA alleles of interest to validate GvL mHA presentation via targeted MS with heavy-labeled standards.
Results
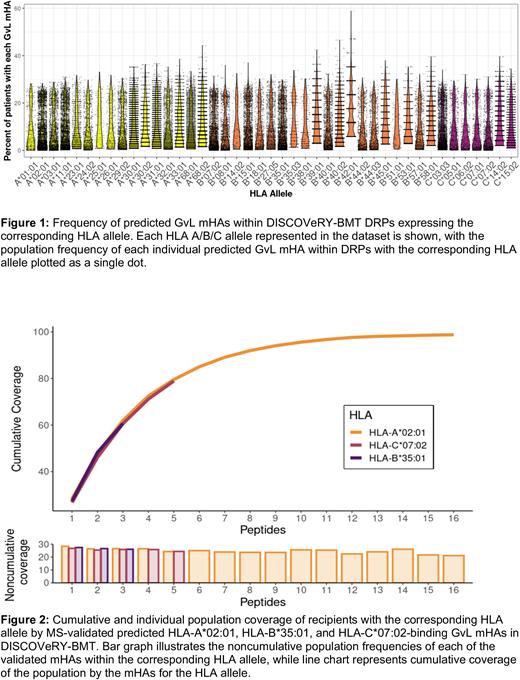
Total number of predicted mHAs and count within each predicted mHA class significantly differed by recipient genomic ancestry, with European American>Hispanic>African American. Number of mHAs also differed markedly by HLA allele. Despite differing total numbers, similar trends in mHA population frequency were observed. While most mHAs are unshared or shared between <10% of patients within an HLA allele, a small population that covers from 20-30% of DRPs is observed in most HLA alleles (Figure 1). Results from the greedy algorithm indicate that for most HLA alleles, only 11-15 GvL mHAs are needed to cover 100% of DRPs in DISCOVeRY-BMT with a given HLA allele.
We then used MS to validate a total of 24 novel predicted shared GvL mHAs for three common HLA alleles. The 16 HLA-A*02:01-binding, 3 HLA-B*35:01-binding, and 5 HLA-C*07:02-binding predicted GvL mHAs are cumulatively found within 98.8%, 60.7%, and 78.9% of DRPs of DISCOVeRY-BMT DRPs with the HLA allele (Figure 2). The GvL mHAs also have individual population frequencies between 21.2-28.3% of DRPs with the corresponding HLA allele (Figure 2).
Conclusions
We report here the discovery of a large set of novel shared GvL mHAs. Most mHAs are unshared, but we observed a small set of mHAs that were found in a large number of recipients within most HLA alleles represented in DISCOVeRY-BMT. This group of mHAs represents the shared mHAs our work targeted for cumulative population coverage analysis and validation.
Our 24 validated predicted GvL mHAs approximately double the previously identified 13 GvL mHAs (Oostvogels et al 2016, Lansford et al 2018, Goulmy et al 2004, Amado-Azevedo et al 2018, Choi et al 2020, Kremer et al 2020, Pont et al 2016). The novel mHAs together cover most of the DRPs with the corresponding HLA alleles and also have high individual population frequencies, indicating their potential benefit as therapeutic targets even if only some could be developed into targeted treatments. Our work demonstrates that shared GvL mHA identification is a feasible and promising technique for expanding mHA-targeting immunotherapeutics to increased numbers of alloHCT recipients.
Disclosures
Pasquini:Novartis: Research Funding; Kite: Research Funding; Bristol Myers Squibb: Consultancy, Research Funding; Janssen: Research Funding. McCarthy:Genentech: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Sanofi: Consultancy; Oncopeptides: Consultancy, Honoraria; Novartis: Consultancy, Honoraria; Takeda Pharmaceuticals America, Inc.: Consultancy, Honoraria; Magenta Therapeutics: Consultancy, Honoraria; Starton Therapeutics: Consultancy, Honoraria; Juno: Consultancy, Honoraria; Fate Therapeutics: Consultancy, Honoraria; Bluebird Bio: Consultancy, Honoraria; Axios: Consultancy, Honoraria; Karyopharm Therapeutics Inc.: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Partner Therapeutics, Inc.: Consultancy, Honoraria; Janssen Global Services, LLC: Consultancy, Honoraria; Bristol Myers Squibb Company: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Sucheston-Campbell:Roche: Current Employment, Current equity holder in publicly-traded company.
Author notes
 This icon denotes a clinically relevant abstract
This icon denotes a clinically relevant abstract
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal